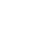Recent Publications
Complete List of Publications
Google Scholar profile , ORCID 0000-0001-9792-5015
†Undergraduate authors, OA Open access
2025
OA Chang SH, George W, DC Nelson DC. (2025) “An N-terminal domain specifies developmental control by the SMAX1-LIKE family of transcriptional regulators in Arabidopsis thaliana.” PNAS USA. 122 (24) e2412793122.
OA Das D, Varshney K, Ogawa S, Torabi S, Hüttl R, Nelson DC, Gutjahr C. (2025) “Ethylene promotes SMAX1 accumulation to inhibit arbuscular mycorrhiza symbiosis.” Nature Communications. 16, 2025.
Chang SH, George W, DC Nelson DC. (2025) “Transcriptional regulation of development by SMAX1-LIKE proteins – targets of strigolactone and karrikin/KAI2 ligand signaling.” Journal of Experimental Botany. eraf027.
*Zhou A, *Kane A, Wu S, Wang K, †Santiago M, Ishiguro Y, Yoneyama K, Palayam M, Shabek N, *Xie X, *Nelson DC, *Li Y. (2025) “Evolution of interorganismal strigolactone biosynthesis in seed plants.” Science. 387 (6731), eadp0779.
2024
White ARF, Kane A, Ogawa S, Shirasu K, Nelson DC. (2024) “Dominant-negative KAI2d paralogs putatively attenuate strigolactone responses in root parasitic plants.” Plant & Cell Physiology.
OA Li Q, Yu H, Chang W, Chang S, Guzmán M, Faure L, Wallner E-S, Yan H, Greb T, Wang L, Yao R, Nelson DC. (2024) “SMXL5 attenuates strigolactone signaling in Arabidopsis thaliana by inhibiting SMXL7 degradation.” Molecular Plant. 17(4):631-647.
2023
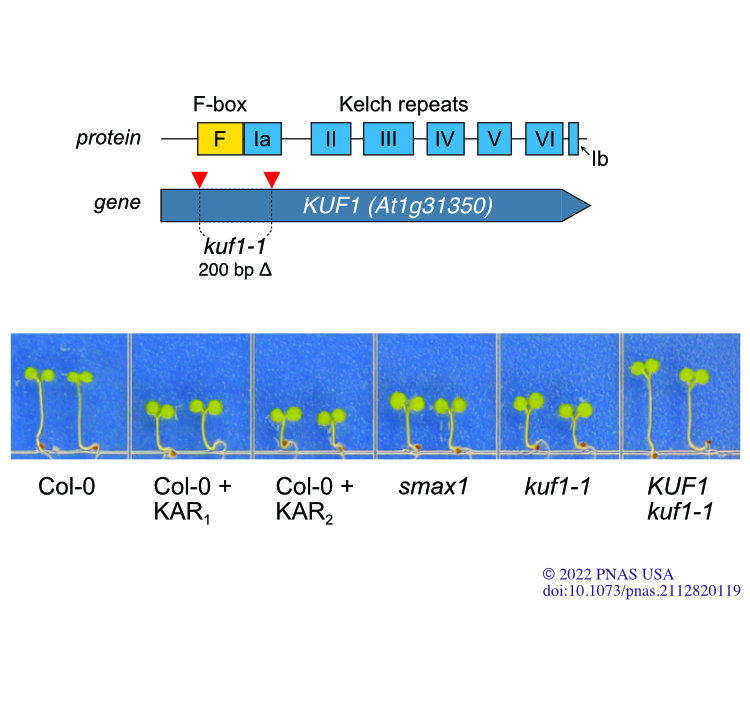
OA Waters MT, Nelson DC. (2023) “Karrikin perception and signalling.” New Phytologist. 237 (5), 1525-1541.
2022
OA Ogawa S, Cui S, White ARF, Nelson DC, Yoshida S, Shirasu K. (2022) “Strigolactones are chemoattractants for host tropism in Orobanchaceae parasitic plants.” Nature Communications. 13(1).
OA †Martinez SE, Conn CE, Guercio AM, Sepulveda C, Fiscus CJ, Koenig D, Shabek N, Nelson DC. (2022) “A KARRIKIN INSENSITIVE2 paralog in lettuce mediates highly sensitive germination responses to karrikinolide.” Plant Physiology. 190(2), 1440-1456.
Ito A, Choi JH, Yokoyama-Maruyama W, Kotajima M, Wu J, Suzuki T, Terashima Y, Suzuki H, Hirai H, Nelson DC, Tsunematsu Y, Watanabe K, Asakawa T, Ouchi H, Inai M, Dohra H, Kawagishi H. (2022) “1,2,3-Triazine formation mechanism of the fairy chemical 2-azahypoxanthine in the fairy ring-forming fungus Lepista sordida.” Organic & Biomolecular Chemistry. 20(13), 2636–2642.
Chen J, Nelson DC, Shukla D. (2022) “Activation Mechanism of Strigolactone Receptors and Its Impact on Ligand Selectivity between Host and Parasitic Plants.” Journal of Chemical Information and Modeling. 62(7), 1712–1722.
White ARF, †Mendez JA, Khosla A, Nelson DC. (2022) “Rapid analysis of strigolactone receptor activity in a Nicotiana benthamiana dwarf14 mutant.” Plant Direct 6(3).
Li Q, Martín-Fontecha ES, Khosla A, White ARF, Chang S, Cubas P, Nelson DC. (2022) “The strigolactone receptor D14 targets SMAX1 for degradation in response to GR24 treatment and osmotic stress.” Plant Communications, 3(2), 100303.
Tian H, Watanabe Y, Nguyen KH, Tran CD, Abdelrahman M, Liang X, Xu K, Sepulveda C, Mostofa MG, Van Ha C, Nelson DC, Mochida K, Tian C, Tanaka M, Seki M, Miao Y, Tran L-SP, Li W. (2022) “KARRIKIN UPREGULATED F-BOX 1 negatively regulates drought tolerance in Arabidopsis.” Plant Physiology.
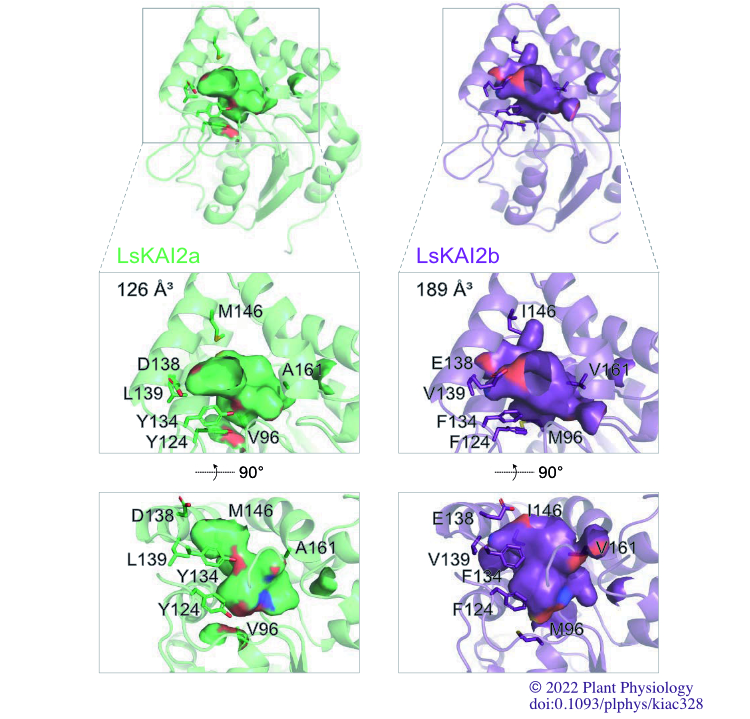
Sepulveda C, Guzmán MA, Li Q, Villaécija-Aguilar JA, UGMartinez SE, Kamran M, Khosla A, Liu W, Gendron JM, Gutjahr C, Waters MT, Nelson DC. (2022) “KARRIKIN UP-REGULATED F-BOX 1 (KUF1) imposes negative feedback regulation of karrikin and KAI2 ligand metabolism in Arabidopsis thaliana.” PNAS USA, 119(11).
2021
Bursch K, Niemann ET, Nelson DC, Johansson H. (2021) “Karrikins control seedling photomorphogenesis and anthocyanin biosynthesis through a HY5-BBX transcriptional module.” The Plant Journal, 107(5), 1346–1362.
Yoda A, Mori N, Akiyama K, Kikuchi M, Xie X, Miura K, Yoneyama K, Sato-Izawa K, Yamaguchi S, Yoneyama K, Nelson DC, Nomura T. (2021) “Strigolactone biosynthesis catalyzed by cytochrome P450 and sulfotransferase in sorghum.” New Phytologist, 232(5), 1999–2010.
Mills A, Jaganatha V, Cortez A, Guzmán M, Burnette JM, Collin M, Lopez-Lopez B, Wessler SR, Van Norman JM, Nelson DC, Rasmussen CG. (2021) “A Course-Based Undergraduate Research Experience in CRISPR-Cas9 Experimental Design to Support Reverse Genetic Studies in Arabidopsis thaliana.” Journal of Microbiology & Biology Education, 22(2).
Chen J, White A, Nelson DC, Shukla D. (2021) “Role of substrate recognition in modulating strigolactone receptor selectivity in witchweed.” Journal of Biological Chemistry, 297(4), 101092.
Nelson DC. (2021) “The mechanism of host-induced germination in root parasitic plants.” Plant Physiology. 185(4), 1353–1373.
Yao J, Scaffidi A, Meng Y, Melville KT, Komatsu A, Khosla A, Nelson DC, Kyozuka J, Flematti GR, Waters MT. (2021) “Desmethyl butenolides are optimal ligands for karrikin receptor proteins.” New Phytologist.
2020
Khosla A, Nelson DC. (2020) “Ratiometric Measurement of Protein Abundance after Transient Expression of a Transgene in Nicotiana benthamiana.” Bio-protocol 10 (17), e3747-e3747.
Khosla A, Morffy N, Li Q, Faure L, Chang SH, Yao J, Zheng J, †Cai ML, Stanga J, Flematti GR, Waters MT, Nelson DC. (2020) “Structure–Function Analysis of SMAX1 Reveals Domains That Mediate Its Karrikin-Induced Proteolysis and Interaction with the Receptor KAI2” The Plant Cell. 32(8):2639-2659.
Khosla A, Rodriguez-Furlan C, †Kapoor S, Van Norman JM, Nelson DC. (2020) “A series of dual-reporter vectors for ratiometric analysis of protein abundance in plants.” Plant Direct. 4(6):e00231
Blázquez MA, Nelson DC, Weijers D. (2020) “Evolution of plant hormone response pathways.” Annual Review of Plant Biology. 71:327-353. (invited review)
2019
Yoshida S, Kim S, Wafula EK, Tanskanen J, Kim Y-M, Honaas L, Yang Z, Spallek T, Conn CE, Ichihashi Y, Cheong K, Cui S, Der JP, Gundlach H, Jiao Y, Hori C, Ishida JK, Kasahara H, Kiba T, Kim M-S, Koo N, Laohavisit A, Lee Y-H, Lumba S, McCourt P, Mortimer JC, Mutuku JM, Nomura T, Sasaki-Sekimoto Y, Seto Y, Wang Y, Wakatake T, Sakakibara H, Demura T, Yamaguchi S, Yoneyama K, Manabe R, Nelson DC, Schulman AH, Timko MP, dePamphilis CW, Choi D, and Shirasu, K. (2019) “Genome Sequence of Striga asiatica Provides Insight into the Evolution of Plant Parasitism.” Current Biology, 29(18), 3041–3052.e4
2017
Li W, Nguyen KH, Chu HD, Ha CV, Watanabe Y, Osakabe Y, Leyva-Gonzalez MA, Sato M, Toyooka K, †Voges L, Tanaka M, Mostofa MG, Seki M, Seo M, Yamaguchi S, Nelson DC, Tian C, Herrera-Estrella L, Tran LP. (2017) “The karrikin receptor KAI2 promotes drought resistance in Arabidopsis thaliana.” PLoS Genetics 13:e1007076
Bythell-Douglas R, Rothfels C, Stevenson D, Graham SW, Wong GK-S, Nelson DC, Bennett T. (2017) “Evolution of strigolactone receptors by gradual neo-functionalization of KAI2 paralogues.” BMC Biology 10.1186/s12915-017-0397-z
Conn CE, Nelson DC. (2017) “It’s not easy being not green: the making of a parasitic plant.” Teaching Tools in Plant Biology: Lecture Notes. The Plant Cell (online)
Waters MT, Gutjahr C, Bennett T, Nelson DC. (2017) “Strigolactone signaling and evolution.” Annual Review of Plant Biology, 68:291-322. Free reprint.
Cutler S, Nelson DC. (2017) “Plant hormones.” in: eLS. John Wiley & Sons Ltd, Chichester.
2016
Khosla A, Nelson DC. (2016) “Strigolactones, super hormones in the fight against Striga.” Current Opinion in Plant Biology, 33:57-6.
Lopez-Obando M, Conn CE, Hoffmann B, Bythell-Douglas R, Nelson DC, Rameau C, Bonhomme S. “Structural modelling and transcriptional responses highlight a clade of PpKAI2-LIKE genes as candidate receptors for strigolactones in Physcomitrella patens.” Planta, 243(6):1441-53.
Morffy N*, Faure L*, Nelson DC. (2016) “Smoke and hormone mirrors: action and evolution of karrikin and strigolactone signaling.” Trends in Genetics, 32(3):176-88.
Stanga JP, Morffy N, Nelson DC. (2016) “Functional redundancy in the control of seedling growth by the karrikin signaling pathway.” Planta, 243(6):1397-1406.
Conn CE, Nelson DC. (2016) “Evidence that KARRIKIN-INSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not karrikin or strigolactone.” Frontiers in Plant Science, 6:1219. Open Access
2015
Soundappan I*, Bennett T*, Morffy N, Yueyang L, Stanga JP, †Abbas A, Leyser O, Nelson DC. (2015) “SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis.” Plant Cell, 27:3143-59.
Conn CE, Bythell-Douglas R, †Neumann D, Yoshida S, †Whittington B, Westwood JH, Shirasu K, Bond CS, Dyer KA, Nelson DC. (2015) “Convergent evolution of strigolactone perception enabled host detection in parasitic plants.” Science, 349:540-43. Free Abstract, Full text, or Reprint.
2013
Stanga JP, Smith SM, Briggs WR, Nelson DC. (2013) “SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis.” Plant Physiology, 163(1):318-330.
Nelson DC “Are karrikin signalling mechanisms relevant to strigolactone perception?” in Parasitic Orobanchaceae, edited by Joel DM, Gressel J, Musselman LJ, 221-230. Springer Berlin Heidelberg, 2013.
2012
Nelson DC, Flematti GR, Ghisalberti EL, Dixon KW, Smith SM. (2012) “Regulation of seed germination and seedling growth by chemical signals from burning vegetation.” Annual Reviews of Plant Biology, 63: 107-130. Free reprint.
Waters MT, Nelson DC, Scaffidi A, Flematti GR, †Sun Y, Dixon KW, Smith SM. (2012) “Specialization within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis.” Development, 139(7):1285-1295.
Additional publications (2011 and prior)
Waters MT, Smith SM, Nelson DC. (2011) “Smoke signals and seed dormancy: Where next for MAX2?” Plant Signaling & Behavior, 6(9):1418-1422.
Nelson DC, Scaffidi A, Dun EA, Waters, MT, Flematti GR, Dixon KW, Beveridge CA, Ghisalberti EL, Smith SM. (2011)
“F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana.” PNAS USA, 108(21):8897-8902.
Scaffidi A, Flematti GR, Nelson DC, Dixon KW, Smith SM, Ghisalberti EL. (2011) “The synthesis and biological evaluation of labelled karrikinolides for the elucidation of the mode of action of the seed germination stimulant.” Tetrahedron, 67(1):152-157.
Purdy SJ, Bussell JD, Nelson DC, Villadsen D, Smith S. (2011) “A nuclear-localized protein, KOLD SENSITIV-1, affects the expression of cold-responsive genes during prolonged chilling in Arabidopsis.” Journal of Plant Physiology, 168(3):263-269.
Nelson DC, Flematti GR, †Riseborough J, Ghisalberti EL, Dixon KW, Smith SM. (2010) “Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana.” PNAS USA, 107(15):7095-7100.
Flematti GR, Scaffidi A, Goddard-Borger ED, Heath CH, Nelson DC, Commander LE, Stick RV, Dixon KW, Smith SM, Ghisalberti EL. (2010) “Structure-activity relationship of karrikin germination stimulants.” Journal of Agricultural and Food Chemistry, 58(15):8612-8617.
Chiwocha SDS, Dixon KW, Flematti GR, Ghisalberti EL, Merritt DJ, Nelson DC, †Riseborough J, Smith SM, Stevens JC. (2009) “Karrikins: A new family of plant growth regulators in smoke.” Plant Science, 177(4):252-256.
Nelson DC, †Riseborough J, Flematti GR, Stevens JC, Ghisalberti EL, Dixon KW, Smith SM. (2009) “Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light.” Plant Physiology, 149(2):863-873.
Pandey S, Nelson DC, Assmann SM. (2009) “Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis.” Cell, 136(1):136-148.
Nelson DC, Wohlbach DJ, Rodesch MR, Stolc V, Sussman MR, Samanta MP. (2007) “Identification of an in vitro transcription-based artifact affecting oligonucleotide microarrays.” FEBS Letters, 581(18):3363-3370.
Stolc V, Samanta MP, Tongprasit W, Sethi H, Liang S, Nelson DC, Hegeman A, Nelson C, Rancour D, Bednarek S, Ulrich EL, Zhao Q, Wrobel RL, Newman CS, Fox BG, Phillips GN, Markley JL, Sussman MR. (2005) “Identification of transcribed sequences in Arabidopsis thaliana by using high-resolution genome tiling arrays.” PNAS USA, 102(12):4453-4458.
Lasswell J, Rogg LE, Nelson DC, Rongey C, Bartel B. (2000) “Cloning and characterization of IAR1, a gene required for auxin conjugate sensitivity in Arabidopsis.” Plant Cell, 12(12):2395-2408.
Nelson DC, Lasswell J, Rogg LE, Cohen MA, Bartel B. (2000) “FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis.” Cell, 101(3):331-340.