Sepulveda C, Guzmán MA, Li Q, Villaécija-Aguilar JA, UGMartinez SE, Kamran M, Khosla A, Liu W, Gendron JM, Gutjahr C, Waters MT, Nelson DC. (2022) “KARRIKIN UP-REGULATED F-BOX 1 (KUF1) imposes negative feedback regulation of karrikin and KAI2 ligand metabolism in Arabidopsis thaliana.” PNAS USA, 119(11).
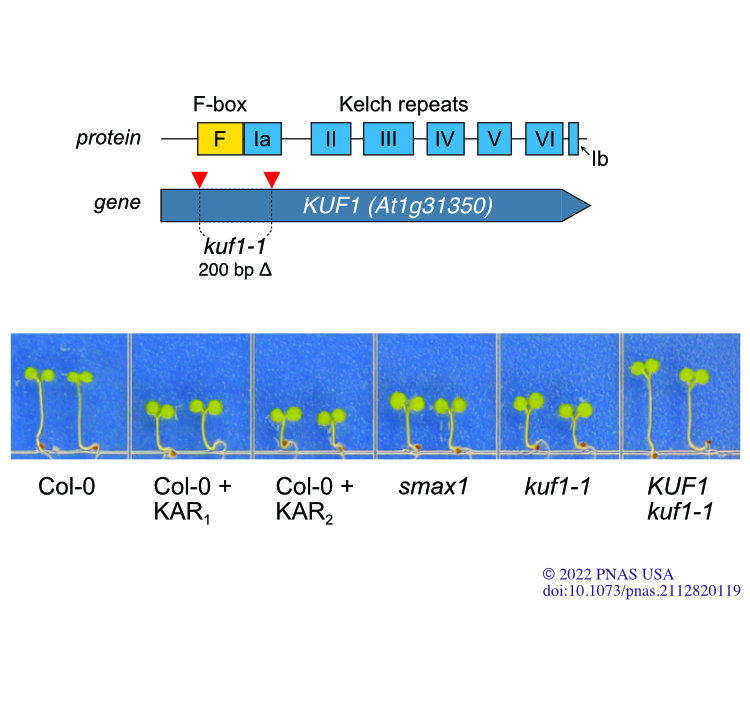
This was a really fun story to put together. It began as a side project for Claudia Sepulveda; the goal was for us to gain some initial experience using CRISPR-Cas9 gene-editing technologies, which we had not tried before. Claudia targeted KUF1, an F-box gene we had been using as a transcriptional marker for KAR/KL responses for several years. Prior experience has made me a little pessimistic about reverse genetic studies, but Claudia found an interesting phenotype: the kuf1 mutant seedlings looked like they were treated with karrikins already! Further, kuf1 phenotypes required a functional KAR/KL signaling pathway to be expressed. This implied that KUF1 functioned as a negative regulator of KAR/KL responses.
While testing how kuf1 seedlings responded to karrikin treatments, Claudia made a critical observation. We had gotten into the habit of considering KAR1 and KAR2 as essentially the same compound. Because KAR2 was more potent we used it in most experiments and often didn’t bother with KAR1. Claudia, however, found that kuf1 mutants were specifically hypersensitive to KAR1 and not KAR2 or another synthetic KAI2 agonist, GR24. This indicated that KUF1 was likely affecting the ability of the KAR/KL signaling pathway to perceive specific compounds, not the activity of the pathway overall. Through a series of experiments, we ultimately concluded that KUF1 affects the conversion of KARs into active signals and KL metabolism. The catch, of course, is that we don’t yet know what these KAR metabolites or KL are, so this conclusion rests on logical arguments. We are currently working on finding these compounds and figuring out what happens downstream of KUF1.