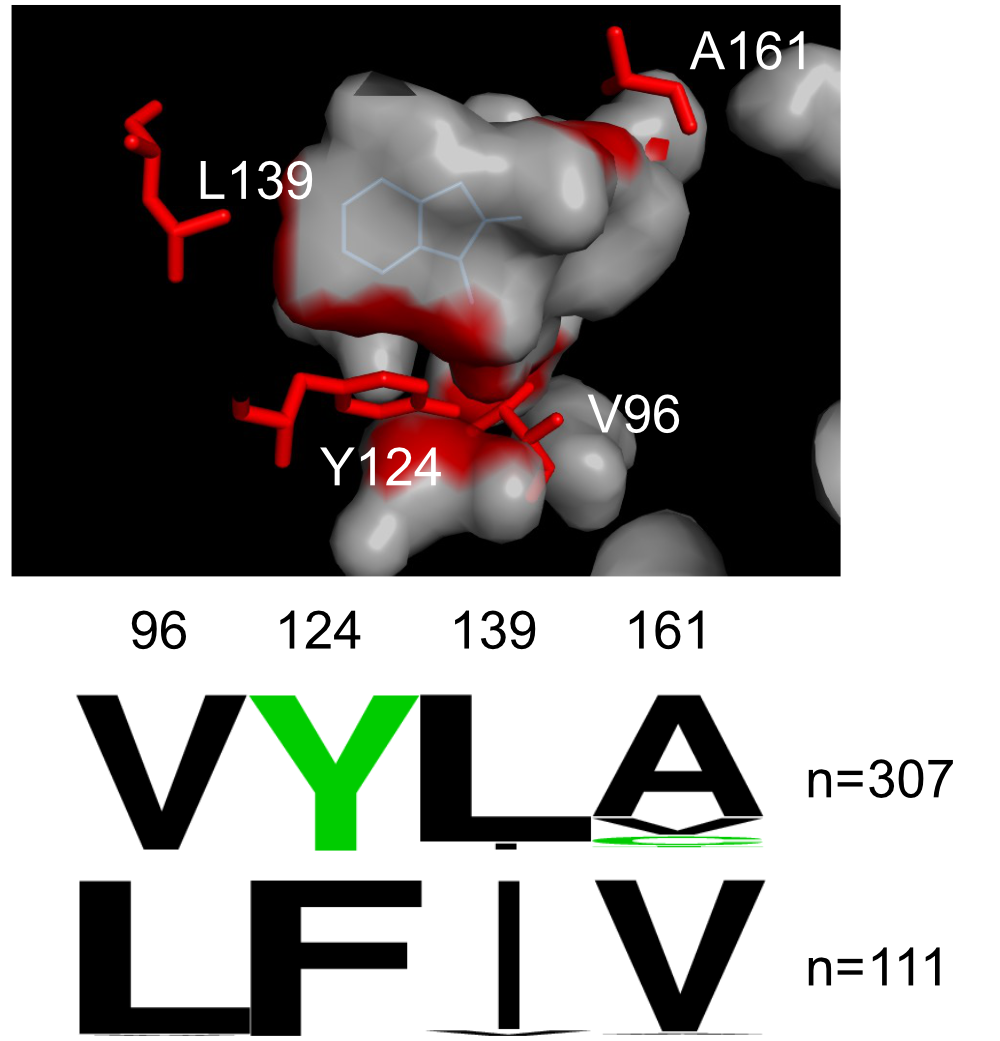
 The highly similar nature of the proteins that carry out karrikin and strigolactone signal transduction raises the question of how signaling fidelity is achieved in these pathways. What amino acid differences in KAI2 and D14 allow these proteins to selectively respond to different compounds? What amino acid substitutions have led to preferred interactions between KAI2 and SMAX1/SMXL2, and D14 with SMXL6, SMXL7, and SMXL8?
The highly similar nature of the proteins that carry out karrikin and strigolactone signal transduction raises the question of how signaling fidelity is achieved in these pathways. What amino acid differences in KAI2 and D14 allow these proteins to selectively respond to different compounds? What amino acid substitutions have led to preferred interactions between KAI2 and SMAX1/SMXL2, and D14 with SMXL6, SMXL7, and SMXL8?
To investigate these questions, we integrate clues from the evolutionary history and protein structures of KAI2 and D14. We then test how mutations of candidate amino acids of interest affect the receptors’ signaling specificity. Ultimately, this knowledge will enable us to engineer cross-wired or orthogonal versions of these signaling pathways, or sensitivity to new molecules, which may lead to new applications in agronomic trait control.
For further reading, see
OA †Martinez SE, Conn CE, Guercio AM, Sepulveda C, Fiscus CJ, Koenig D, Shabek N, Nelson DC. (2022) “A KARRIKIN INSENSITIVE2 paralog in lettuce mediates highly sensitive germination responses to karrikinolide.” Plant Physiology. 190(2), 1440-1456.
Li Q, Martín-Fontecha ES, Khosla A, White ARF, Chang S, Cubas P, Nelson DC. (2022) “The strigolactone receptor D14 targets SMAX1 for degradation in response to GR24 treatment and osmotic stress.” Plant Communications, 3(2), 100303.
Khosla A, Morffy N, Li Q, Faure L, Chang SH, Yao J, Zheng J, †Cai ML, Stanga J, Flematti GR, Waters MT, Nelson DC. (2020) “Structure–Function Analysis of SMAX1 Reveals Domains That Mediate Its Karrikin-Induced Proteolysis and Interaction with the Receptor KAI2” The Plant Cell. 32(8):2639-2659.